At the Forefront of Oncology and Automated Liquid Biopsy

Posted by:
Michael Collasius
Published on:
Apr 15, 2024
Liquid biopsy offers huge potential to transform cancer diagnosis, patient stratification, and treatment monitoring. By improving cancer screening and early detection, liquid biopsy may enable earlier diagnosis and treatment and better outcomes. Its minimally invasive approach, ability to capture tumor heterogeneity, and real-time monitoring make it a powerful tool for personalized treatment strategies.
Challenges and promise of liquid biopsy in oncology
Liquid biopsy – a non-invasive method for detection of circulating free nucleic acids (cfDNA), exosomes, and circulating tumor cells (CTCs) from blood and other biofluids – holds promise for early detection of cancer and treatment monitoring. While tissue biopsy is the current gold standard for detection of cancer, it has many disadvantages. It is invasive and costly, the tumor may not be easily accessible, and it potentially provides only a snapshot of the tumor profile – that may be detected too late. This means that clinicians may not have sufficient information to determine the optimal treatment for patients. Multiple tissue biopsies are associated with increased cost and risks to patients and are not always feasible.
Liquid biopsy overcomes many of the challenges associated with traditional biopsies. This was successfully demonstrated in testing of clinically available BCR::ABL in chronic myelogenous leukemia1 and potentially, in gliomas, for EGFRvIII2 and TERT3.
However, results from a recent study of patients with heavily pretreated, advanced solid tumors demonstrated that there was no association between either baseline circulating tumor DNA (ctDNA) levels or ctDNA decline and patients’ radiographic response or survival4. Its possible that this was due to the heavily treated population or the therapy given. In trials for earlier-stage cancer, ctDNA detection and monitoring is being used to guide treatment decisions, such as whether to give patients adjuvant treatment or switch them from one drug onto another. It provides information on response and progression more quickly than radiologic imaging.
Looking to the future, higher throughputs are needed for liquid biopsy to be used in routine screening for early detection and monitoring of cancer. Wide adoption of liquid biopsy in clinical settings is hindered by a lack of standardized methods. To overcome this, clinical practice guidelines are required for standardization of pre-analytical workflows and improved analytical performance of cfDNA testing.
We are pioneers in automation of life sciences and diagnostics workflows
We specialize in the design and implementation of cutting-edge automated systems for diagnostic and life science companies. Our world-class engineers and scientists understand the unique requirements and complex nature of biological and medical applications and develop automated systems according to ISO 13485.
Some of the automated systems we have successfully developed include:
- Tube handling and capping/decapping
- Sample prep using magnetic beads or spin columns
- Quality control using capillary electrophoresis or optical analysis
- NGS and qPCR analysis
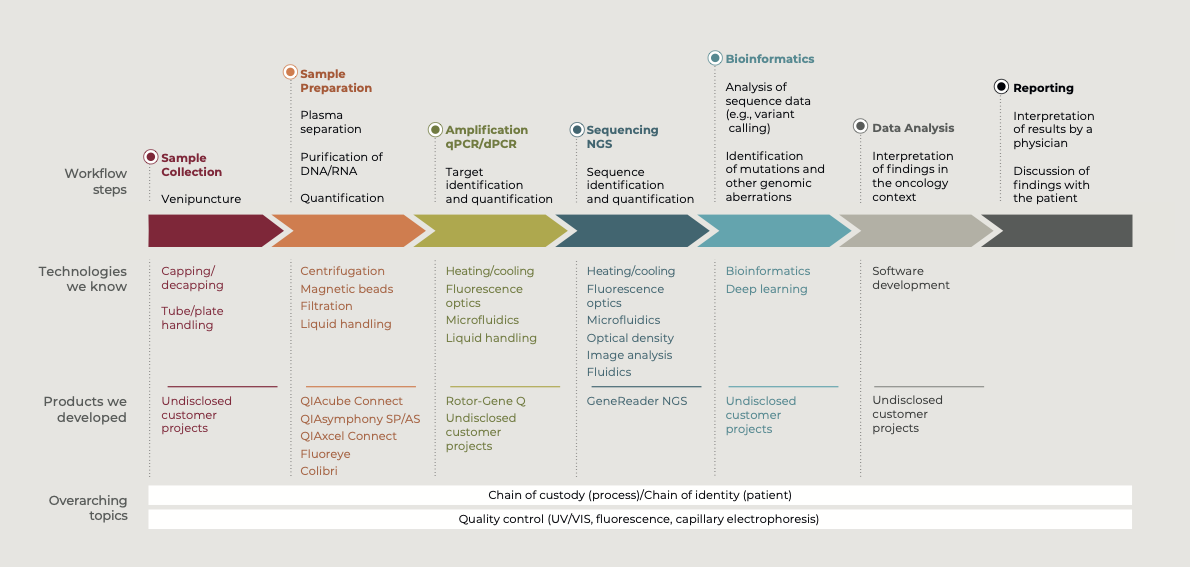
Automated sample prep for challenging samples
Sample prep is one of the most important steps in the liquid biopsy workflow, with increasing demands being placed on the performance of sample prep systems. Due to the low analyte content per mL of sample, liquid biopsy sample prep requires high-volume samples to deliver sufficient yields of cfDNA for downstream analysis. In addition, a specific extraction step for low-molecular-weight nucleic acids is needed to reduce contamination with genomic DNA (i.e., all white blood cells).
We developed the QIAsymphony SP/AS platform for fully automated nucleic acid extraction from 1–96 samples using magnetic beads. To prevent aggregation of the magnetic beads, we have several product concepts to keep the beads uniformly suspended, ensuring reproducible nucleic acid extraction. We developed the QIAcube Connect for extraction of DNA, RNA, and proteins from up to 12 samples using spin columns. These platforms are fully automated for a range of volumes from 1–10 mL and have the required flexibility for liquid biopsy samples.
Traceability of samples is critical to ensure that patients get the right result. The automated sample prep systems we have developed provide a chain-of-custody record that complies with the FDA 21 CFR Part 11 regulations.
Stringent quality control using capillary electrophoresis
Capillary electrophoresis is the method of choice for quality control of cfDNA. It enables fast and easy determination of sample concentration, purity, and size distribution from minimal input volumes. The size distribution shows whether there is contamination with high-molecular-weight DNA. Stringent quality control using capillary electrophoresis means only samples of the highest quality are processed in downstream analyses.
We developed the QIAxcel Connect – fully automated high-resolution DNA or RNA gel electrophoresis from up to 96 samples – for quality control of nucleic acids.
Quality control measurements using optical analysis
The success of next-generation sequencing (NGS) applications depends greatly on sample quality. Only if the concentration and purity are reliably determined can PCR or NGS applications deliver reliable results. We developed the eviDense UV measurement module as an OEM component for manufacturers of PCR and NGS solutions. The intelligent sample handling integrates measurement of concentration and purity of nucleic acid samples into liquid handling workflows. The compact module requires only one microplate location, can analyze 4 wavelengths, and consumes no sample material.
We also developed Fluoreye for Hamilton Robotics. This is a fluorescence detector that can be picked up by a liquid handling probe and moved to the sample to determine nucleic acid concentration and sample quality directly during the pipetting process e.g., for normalizing or pooling samples during NGS library preparation.
Analysis with a fully integrated NGS workflow
The capacity to sequence all 3.2 billion bases of the human genome has increased from 1.3 human genomes sequenced annually to 18,000 human genomes a year within a decade. Despite this improvement, sequencing a genome is still an extremely complex procedure, which prevents its broad use in routine human diagnostics.
We developed the GeneReader NGS – the only integrated NGS workflow from sample to insight. The complete NGS workflow with seamlessly integrated automated components offers ease of use and efficiency, from sample to result. Validated gene panels and fully integrated bioinformatics deliver actionable insights.
References
1. Ross T.S., Mgbemena V.E. Re-evaluating the role of BCR/ABL in chronic myelogenous leukemia. Mol. Cell. Oncol. 2014;1:e963450.
2. Zhang X., Zhao W., Wei W., You Z., Ou X., Sun M., Yin Y., Tang X., Zhao Z., Hu C., et al. Parallel Analyses of Somatic Mutations in Plasma Circulating Tumor DNA (ctDNA) and Matched Tumor Tissues in Early-Stage Breast Cancer. Clin. Cancer Res. 2019;25:6546–6553.
3. Vasseur A., Kiavue N., Bidard F.-C., Pierga J.-Y., Cabel L. Clinical utility of circulating tumor cells: an update. Mol. Oncol. 2021;15:1647–1666.
4. Hu Y, Narayan A, Xu Y, Wolfe J, Vu D, Trinh T, Kantak C, Ivy SP, Eder JP, Deng Y, LoRusso P, Kim JW, Patel AA. Circulating Tumor DNA Dynamics Fail to Predict Efficacy of Poly(ADP-ribose) Polymerase/VEGFR Inhibition in Patients With Heavily Pretreated Advanced Solid Tumors. JCO Precis Oncol. 2024 Feb;8:e2300289. doi: 10.1200/PO.23.00289.
